The SWAN project (Semantic Web Applications in Neuromedicine) developed a practical, common, semantically-structured, framework for scientific discourse initially applied, but not limited, to significant problems in Alzheimer Disease (AD) research. SWAN sets itself the goal to model the scientific discourse about AD and its supporting evidence in a rich way that is compatible with functioning of the current social network as a technology-mediated ecosystem. The SWAN project was the result of a collaboration between the Alzheimer Research Forum (Alzforum) and the MIND Informatics group at Massachusetts General Hospital.
SWAN authoring and publishing process
The SWAN knowledge base is encoded according to the model dictated by the SWAN ontology, such model is used internally by the software platform and does not impact the curators activity directly. The AlzSWAN curators use the web application named Workbench to author the content that will be made available to the large audience through the SWAN Browser, yet a web application.
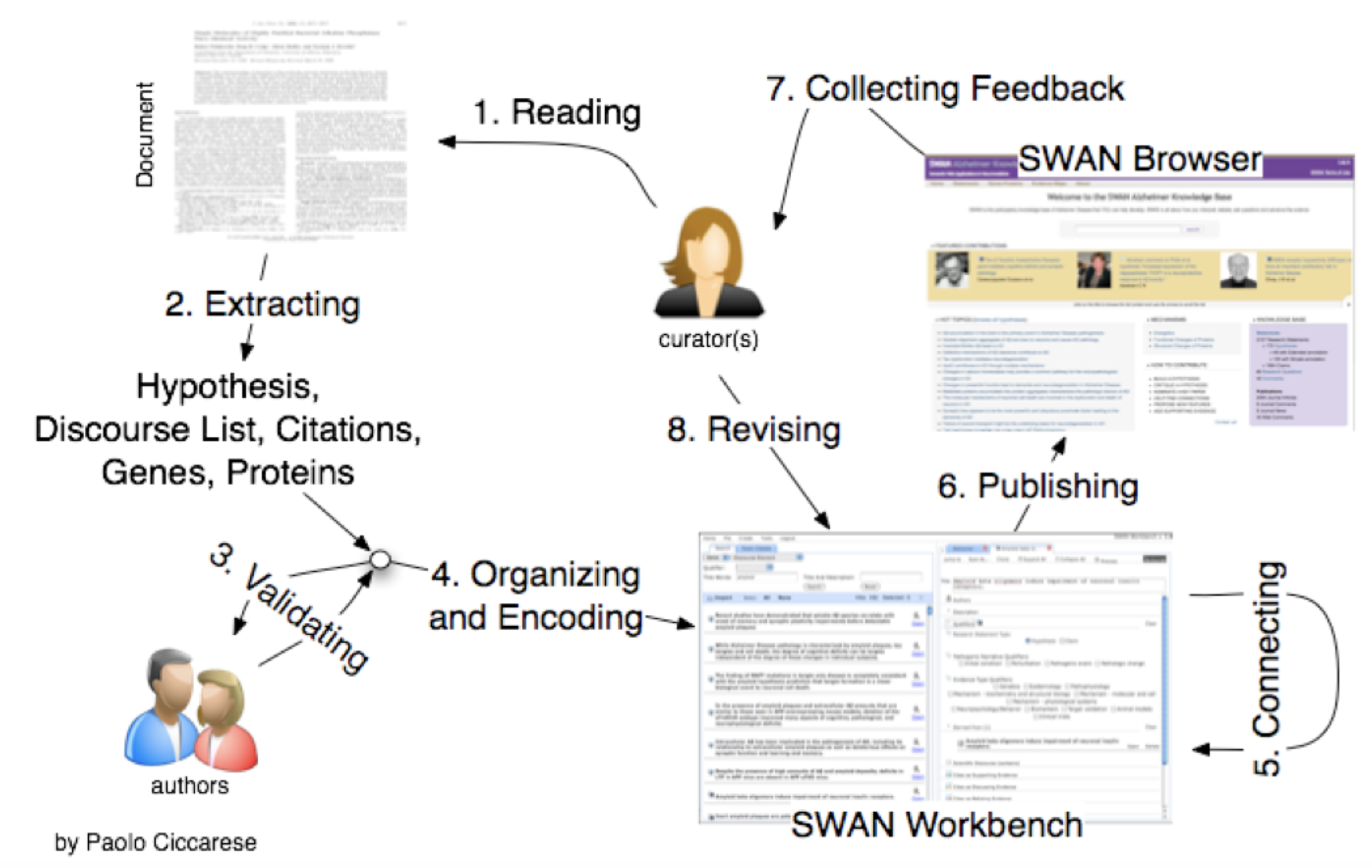
There are different ways the knowledge in AlzSWAN can gets published but the most common case is made of five different steps:
1 The curator is proof reading an article usually representing a hypothesis on AD.
2 The output of the reading process is a linear representation of the scientific discourse of the article which consists of discourse elements: claims, hypotheses and questions.
3 When possible, the representation of the scientific discourse is validated with the authors of the article.
4 For each of the discourse element the curator is selecting related publications, proteins, genes, pathogenic narrative* and evidence types. This knowledge map can be entered through the Workbench a web application to encode knowledge according to the SWAN ontology. In the AlzSWAN implementation, the Workbench is used only by a very limited number of curators, working for AlzForum. Their role is to guarantee correctness and consistency across the whole knowledge base.
5 Through the Workbench is also possible to connect the discourse elements just created with other already existing in the knowledge base.
6 It is possible to save the new content as draft and eventually to publish it in the SWAN Browser developed for the final users. In the case of AlzSWAN the target audience is the community of researchers on AD and neurodegenerative diseases in general.
7 Throught the SWAN Browser it is possible to collect users' feedback through comments.
8 The comments can contribute to refine the scientific discourse representation.
* The pathogenic narrative is a simple, intuitive set of terms that biomedical researchers can use to classify research statements in the context of the Alzheimer disease pathogenic process. The experimental approach ontology provides a simple framework for classifying complementary types of experimental data.
SWAN and the Semantic Web
SWAN has been conceived as a platform for creating, sharing and integrating scientific biomedical knowledge. As the project acronym indicates, one of the initial designing choice has been to adopt technologies proper of the Semantic Web.
The Semantic Web is an extension of the Web through standards by the World Wide Web Consortium (W3C). The standards promote common data formats and exchange protocols on the Web, most fundamentally the Resource Description Framework (RDF).
According to the W3C, "The Semantic Web provides a common framework that allows data to be shared and reused across application, enterprise, and community boundaries". The term was coined by Tim Berners-Lee for a web of data that can be processed by machines. While its critics have questioned its feasibility, proponents argue that applications in industry, biology and human sciences research have already proven the validity of the original concept.
Every piece of information in SWAN is encoded in RDF shaped according to the SWAN Ontology. This allows us not only to elicit the knowledge embedded in scientific articles and scientific communications but also to integrate it with a set of external resources covering biomedical aspects of interest. The new enriched content is then made available for feeding new knowledge integration processes.
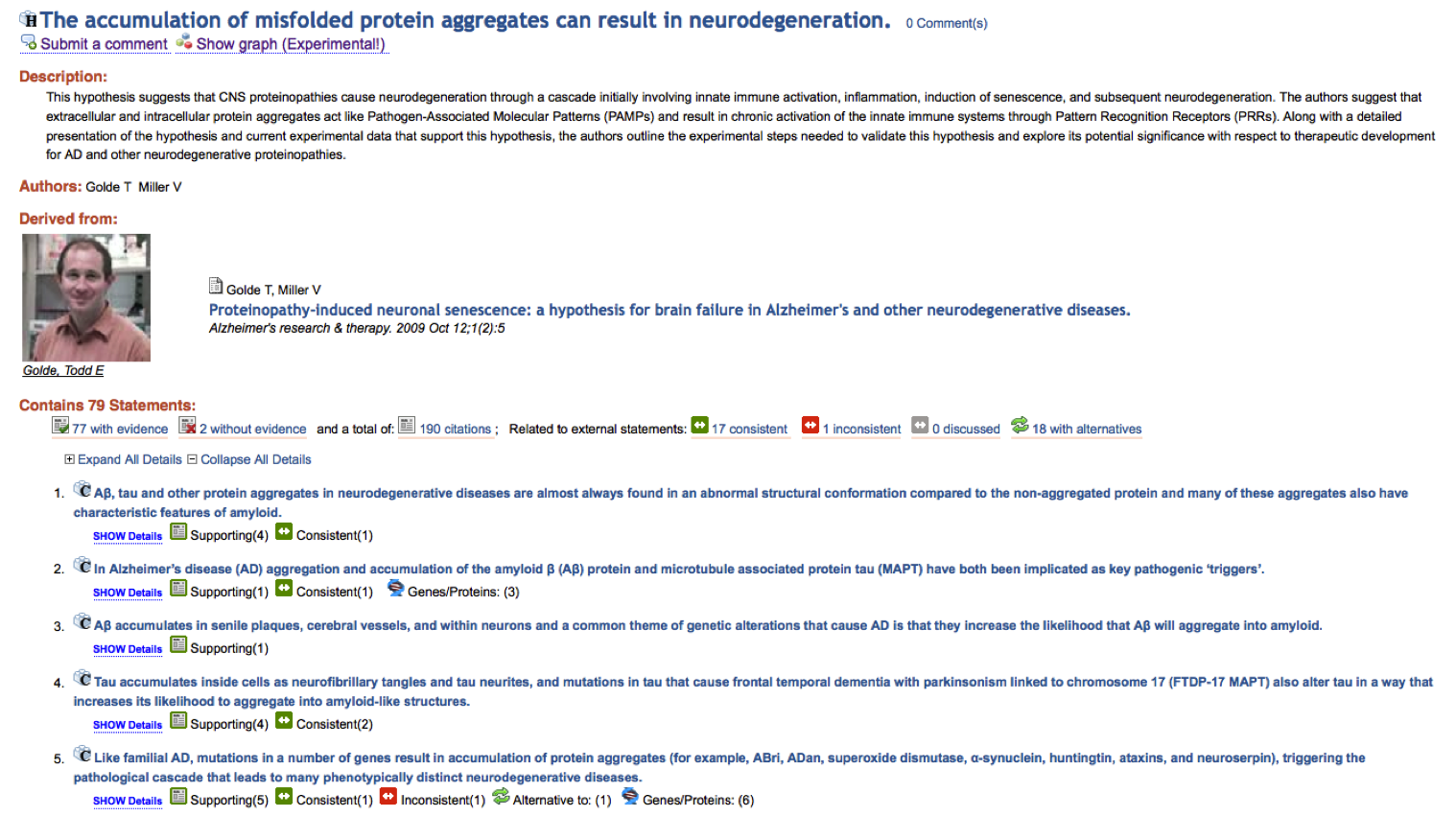
As knowledge is encoded by a RDF network, the SWAN HTML pages like the one (partially represented) above are really just a linear rendering of the knowledge network that is more naturally represented by the following visualization:
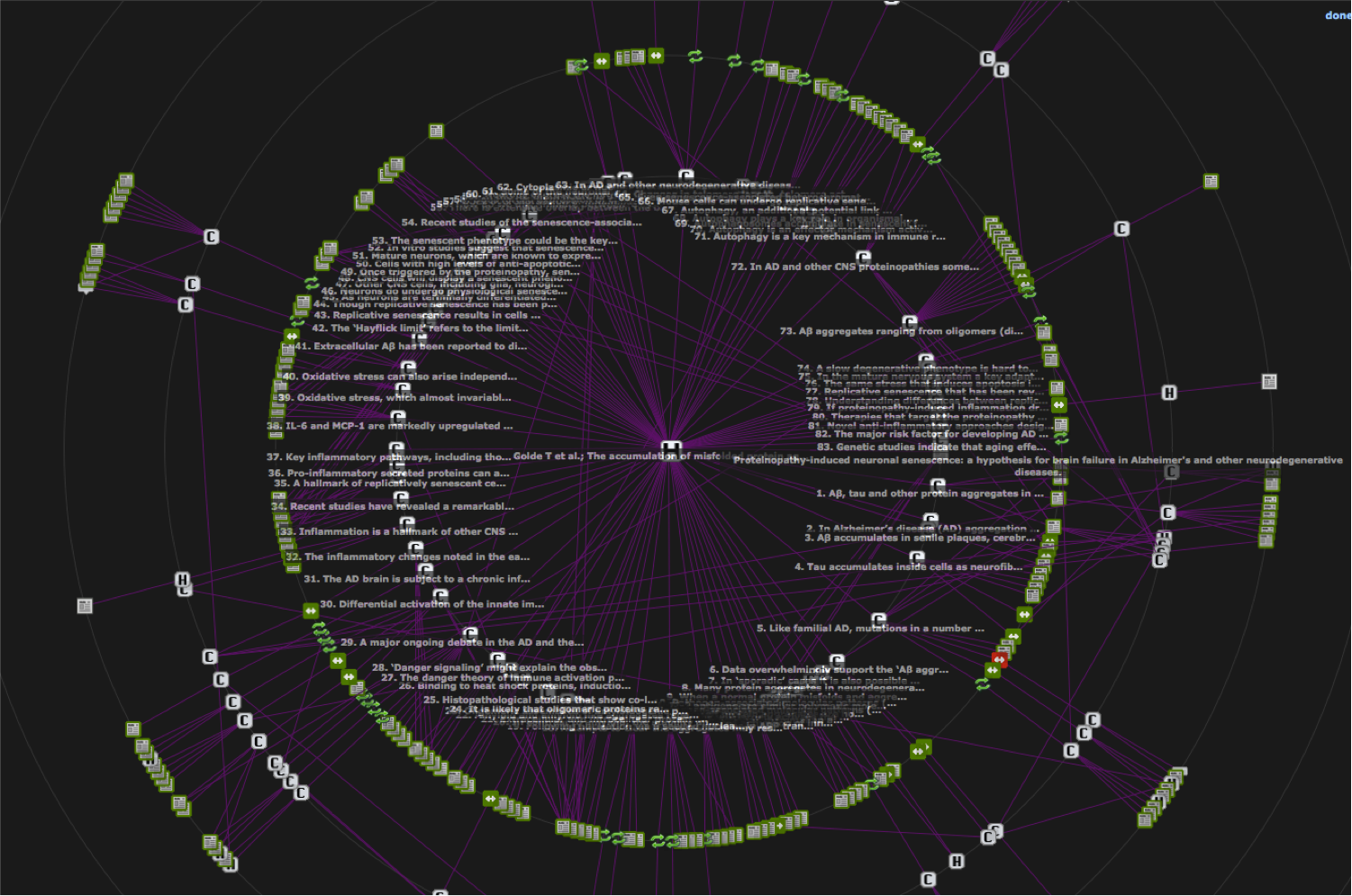
* I've created this interactinve visualization tool for SWAN by customizing one of the graphs offered by the JavaScript InfoVis Toolkit (JIT)
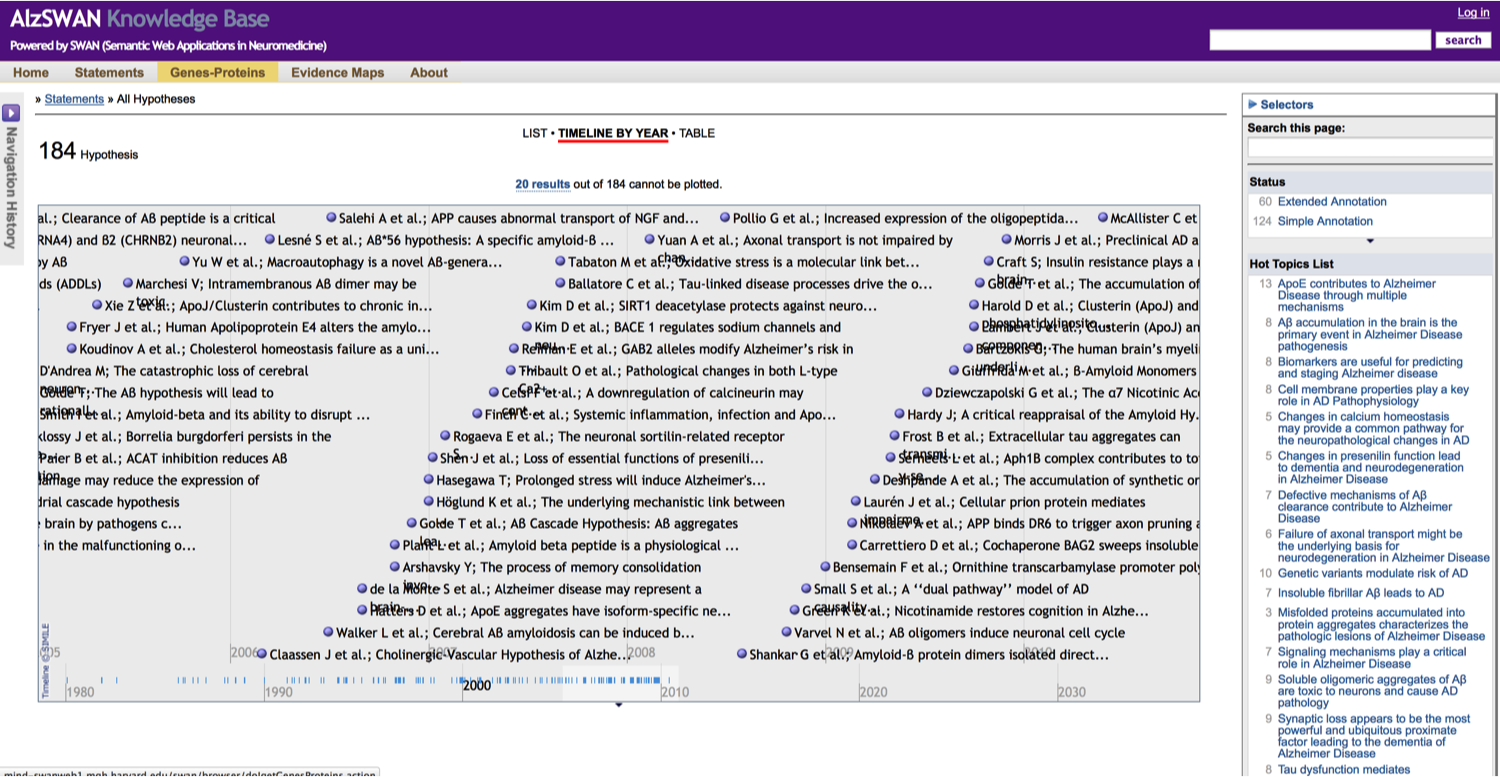
All the hypotheses and claims that have been modeled in AlsSWAN are also available for search. Search is build on a Lucene index that indexes the graphs in several ways each of which pivots on the main discourse element entities: hypotheses, claims and research questions.
Another option for browsing the discourse elements is through the Simile Exhibit and the Simile Timeline: